MCP (Model Context Protocol) server for biothings
Documentation
biothings-mcp
MCP (Model Context Protocol) server for Biothings.io
This server implements the Model Context Protocol (MCP) for BioThings, providing a standardized interface for accessing and manipulating biomedical data. MCP enables AI assistants and agents to access specialized biomedical knowledge through structured interfaces to authoritative data sources. Supported BioThings data sources include:
- mygene.info — Gene annotation and query service
- myvariant.info — Variant annotation and query service
- mychem.info — Chemical compound annotation and query service
If you want to understand more what is Model Context Protocol and how to use it more efficiently you can take DeepLearning AI Course or just search for MCP videos on YouTube.
About MCP (Model Context Protocol)
MCP is a protocol that bridges the gap between AI systems and specialized domain knowledge. It enables:
- Structured Access: Direct connection to authoritative biomedical data sources
- Natural Language Queries: Simplified interaction with specialized databases
- Type Safety: Strong typing and validation through biothings-typed-client
- AI Integration: Seamless integration with AI assistants and agents
Available API Interfaces
This server provides dedicated API interfaces for different BioThings data types, leveraging the biothings-typed-client library. These interfaces are implemented using the following tool handlers:
- Gene Interface:
GeneTools(wrapsGeneClientAsync) - Variant Interface:
VariantTools(wrapsVariantClientAsync) - Chemical Interface:
ChemTools(wrapsChemClientAsync) - Taxon Interface:
TaxonTools(wrapsTaxonClientAsync) - Download Interface:
DownloadTools(provides file download and sequence analysis capabilities)
Local File Saving Features
The server includes local file saving capabilities through the DownloadTools interface, which provides:
Download Tools
- **
download_entrez_data**: Download data from NCBI Entrez databases (returns content as string) - **
download_entrez_data_local**: Download data from NCBI Entrez databases and save to local file
Output Directory Management
- Default Location: Files are saved to
biothings_output/directory in the current working directory - Custom Location: Use
--output-dirparameter to specify a custom output directory - Automatic Creation: Output directories are created automatically if they don't exist
- Unique Filenames: Auto-generated filenames include UUID prefixes to avoid conflicts
Supported File Formats
- FASTA:
.fastaextension for sequence data - GenBank:
.gbextension for GenBank format data - Alignment:
.alnextension for alignment results - JSON:
.jsonextension for structured data - Text:
.txtextension for general text data
Quick Start
Installing uv
# Download and install uv
curl -LsSf https://astral.sh/uv/install.sh | sh
# Verify installation
uv --version
uvx --versionuvx is a very nice tool that can run a python package installing it if needed.
Running with uvx
You can run the biothings-mcp server directly using uvx without cloning the repository:
STDIO Mode (for MCP clients that require stdio, can be useful when you want to save files)
# Run the server in STDIO mode (default mode)
uvx biothings-mcp
# Or explicitly specify stdio mode
uvx --from biothings-mcp stdio
# With custom output directory
uvx --from biothings-mcp stdio --output-dir ./my_dataHTTP Streamable Mode (Web Server)
# Run the server in streamable HTTP mode on default port (3001)
uvx --from biothings-mcp server run
# Run on a custom port
uvx --from biothings-mcp server run --port 8000
# Run on custom host and port
uvx --from biothings-mcp server run --host 0.0.0.0 --port 8000
# With custom output directory
uvx --from biothings-mcp server run --output-dir ./my_dataSSE Mode (Server-Sent Events)
# Run the server in SSE mode on default port (3001)
uvx --from biothings-mcp sse
# Run on a custom port
uvx --from biothings-mcp sse --port 8000The HTTP streamable mode will start a web server that you can access at http://localhost:3001/mcp (with documentation at http://localhost:3001/docs). The STDIO mode is designed for MCP clients that communicate via standard input/output, while SSE mode uses Server-Sent Events for real-time communication.
Configuring your (Anthropic Claude Desktop, Cursor, Windsurf, etc.)
We provide stdio configuration using the proxy (might need npx to run):
- mcp-config-remote.json - for remote configuration
- mcp-config-stdio.json - stdio configuration for localhost for MCP clients which do not support
Inspecting Biothings MCP server
If you want to inspect the methods provided by the MCP use npx (you may need to install nodejs and npm)
Test your MCP setup with the MCP Inspector.
If you want to inspect local streamable-http server you use:
npx @modelcontextprotocol/inspector --config mcp-config.json --server biothings-mcpAdd -remote suffix for the remote server.
If you want to inspect stdio local server you should use
npx @modelcontextprotocol/inspector --config mcp-config-stdio.json --server biothings-mcpYou can also run inspector manually and put server parameters in the interface:
npx @modelcontextprotocol/inspectorAfter that you can explore its methods with MCP Inspector at http://127.0.0.1:6274
Repository setup
# Clone the repository
git clone https://github.com/longevity-genie/biothings-mcp.git
cd biothings-mcp
uv syncRunning the MCP Server
If you already cloned the repo you can run the server with uv
# Start the MCP server in HTTP streamable mode (default port 3001)
uv run server run
# Run in STDIO mode
uv run stdio
# Run in SSE mode
uv run sse
# Run with custom port
uv run server run --port 8000
# Run with custom output directory
uv run server run --output-dir ./my_dataIntegration with AI Systems
To integrate this server with your MCP-compatible AI client, you can use one of the preconfigured JSON files provided in this repository:
- For connecting to a locally running server: Use
mcp-config.json. Ensure the server is running first, either viauv run server(see Running the MCP Server) ordocker-compose up(see Docker Deployment). - For connecting to the publicly hosted server: Use
mcp-config-remote.json. This connects tohttps://biothings.longevity-genie.info/mcpand doesn't require you to run anything locally.
Simply point your AI client (like Cursor, Windserve, ClaudeDesktop, VS Code with Copilot, or others) to use the appropriate configuration file.
Here's an example of how the tools might appear in an MCP client like Cursor after configuration:
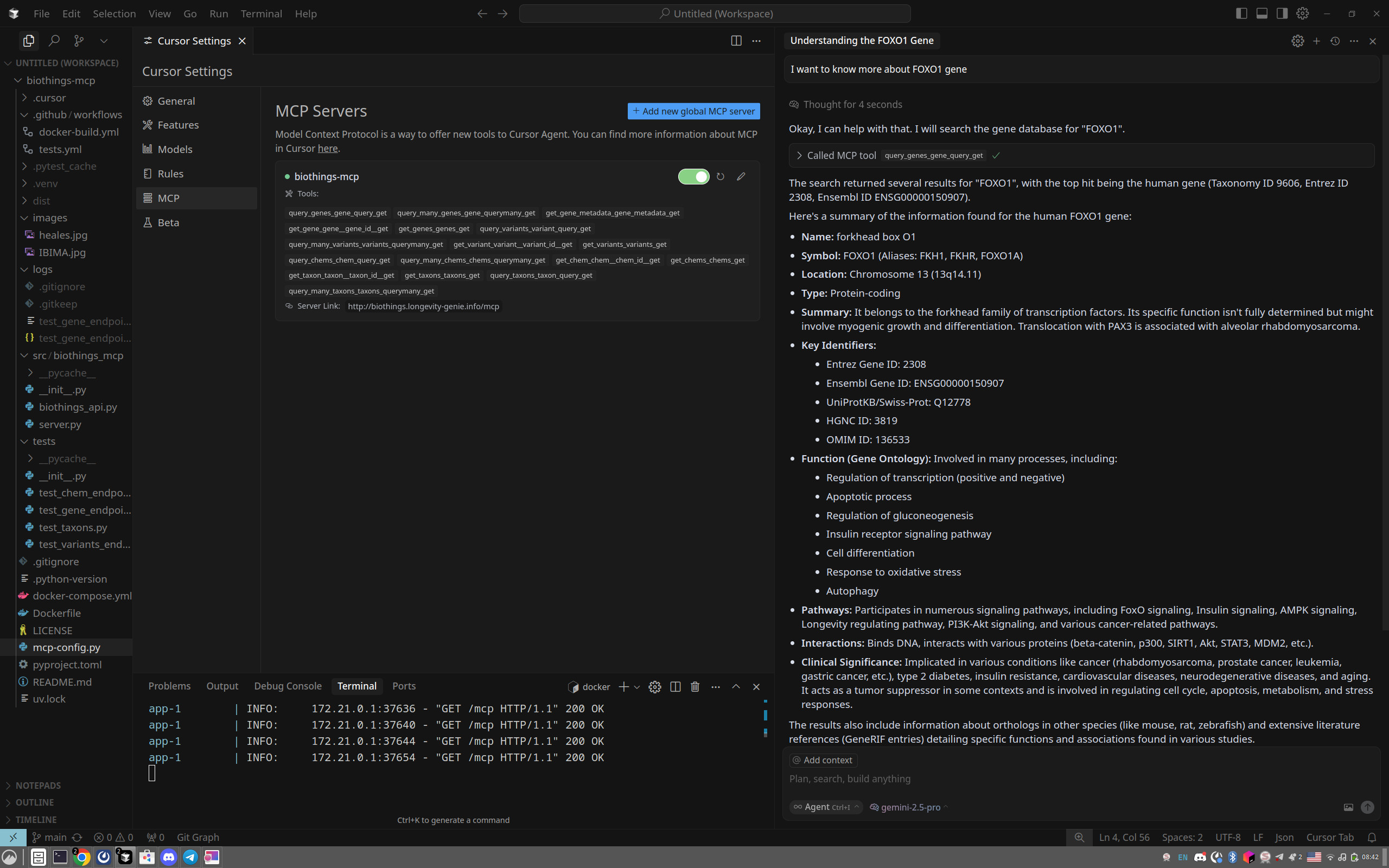
KNOWN ISSUES
The library is beta-quality. The major problem right now is that LLM-s are often stupid and do not know how to put valid gene and gene variant symbols. We plan to mitigrate it by extending comments and providing additional method for entity resolution.
Testing & Verification
Run tests for the API endpoint:
uv run pytest -vvv -sYou can use MCP inspector with locally build MCP server same way as with uvx
*Note: Using the MCP Inspector is optional. Most MCP clients (like Cursor, Windsurv, etc.) will automatically display the available tools from this server once configured. However, the Inspector can be useful for detailed testing and exploration.*
*If you choose to use the Inspector via npx, ensure you have Node.js and npm installed. Using nvm (Node Version Manager) is recommended for managing Node.js versions.*
This opens a web interface where you can explore and test all available tools.
Documentation
For detailed documentation about the MCP protocol and its implementation, refer to:
License
This project is licensed under the MIT License.
Acknowledgments
- BioThings for the REST API and original client library
- MCP Protocol for the protocol specification
- Pydantic for the data validation framework
- FastAPI-MCP for the MCP server implementation
- This project is part of the Longevity Genie organization, which develops open-source AI assistants and libraries for health, genetics, and longevity research.
We are supported by:
*HEALES - Healthy Life Extension Society*
and
IBIMA - Institute for Biostatistics and Informatics in Medicine and Ageing Research
Similar MCP
Based on tags & features
Trending MCP
Most active this week